Heatmap 2D quantification
Heatmaps illustrating plant cell-to-cell connectivity
Dr. Tina Schreier, formerly Prof. Julian Hibberd’s group, Plant Sciences, University of Cambridge; New Phytologist: https://doi.org/10.1111/nph.19343.
This study used 2D- and 3D-electron microscopy to investigate leaf anatomical features of C4 versus C3 plants, providing mechanistic insights on the formation of cell-to-cell connectivity in plants. Plant cells are physically connected through small channels called plasmodesmata (50-100 nm in diameter). The authors found that the formation of these plasmodesmata in the C4 plant seems to be wired to the induction of photosynthesis – the stage at which efficient metabolite transfer through an enhanced cell-to-cell connectivity becomes crucial for photosynthetic efficiency.

C4 photosynthesis is a more efficient type of photosynthesis that evolved in some plants. It entails a compartmentalization of photosynthesis between two different cell types within the leaf - the mesophyll and the bundle sheath cells - to concentrate CO2 around the carbon fixing enzyme RuBisCO. This is in contrast to C3 plants where photosynthesis solely takes place in the mesophyll cells. The abundance of plasmodesmata within a certain cell interface is an indicator of connectivity between the two cells. C4 plants rely heavily on efficient metabolite exchange between mesophyll and bundle sheath cells and are therefore thought to have increased plasmodesmal frequency at this cell interface. Numbers of plasmodesmata in leaves of C4 species Gynandropsis gynandra and its closely related C3 species (Arabidopsis thaliana and Tarenaya hassleriana) were assessed using two different electron microscopy approaches: Large 2D stitched SEM image maps and 3D serial blockface SEM imaging.
At CAIC, leaves of these C4 and C3 plant species were fixed, stained and resin embedded for EM and leaf cross sections were mounted for 2D SEM blockface imaging. Low magnification overviews of the sections allowed to pinpoint the exact areas of interest for quantification.
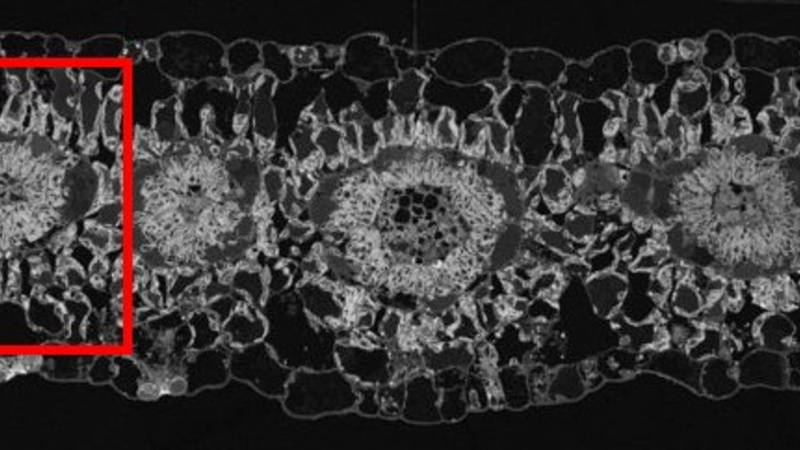
SEM blockface overview of C4 plant Gynandropsis gynandra leaf cross section. The red square highlights a vein cross section depicting the typical C4 leaf Kranz anatomy, the area of interest for plasmodesmata quantification between mesophyll-mesophyll, mesophyll-bundle sheath and bundle sheath-bundle sheath cells.
Automated image acquisition via MAPS software was used to prepare high resolution image maps of the chosen ROI, allowing for the quantification of plasmodesmata.
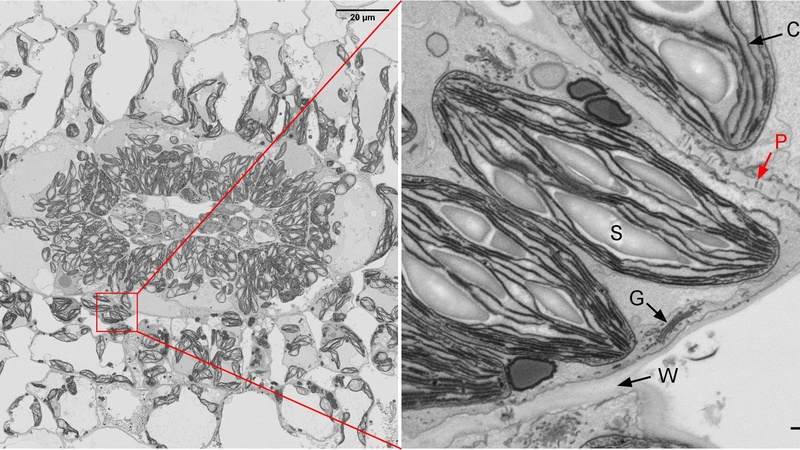
High resolution stitched image map of a vein cross section (left); zooming in on any area within the HR map reveals the cellular structures in more detail (right): C = chloroplast, S = starch granule, W = cell wall, G = Golgi; the red arrow denotes a plasmodesmata crossing the cell wall.
Counting the number of plasmodesmata per mm cell wall between the different leaf cell interfaces allowed the generation of coloured 2D heatmaps of plasmodesmata distribution, where the connectivity between the various cell interfaces in the leaf cross section is immediately obvious.
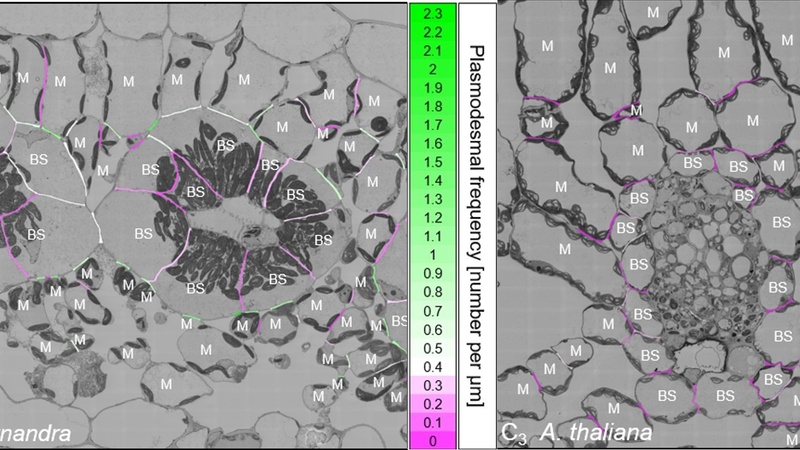
Heatmaps of G. gynandra (C4, left) and A. thaliana (C3, right) vein cross sections where cell connectivity is labelled from low (purple) to high (green); BS = bundle sheath cells, M = mesophyll cells.
The result of plasmodesmata quantification via large 2D stitched maps performed at CAIC was compared to that obtained by 3D serial blockface imaging performed at the Institute of Molecular Plant Biology at the ETH Zürich. This comparison showed both methods gave consistent results, with 2D map counting allowing for higher throughput/counting of a much larger number of cell interfaces, whilst SBF-SEM allowed for image segmentation and reconstruction with excellent 3D views.
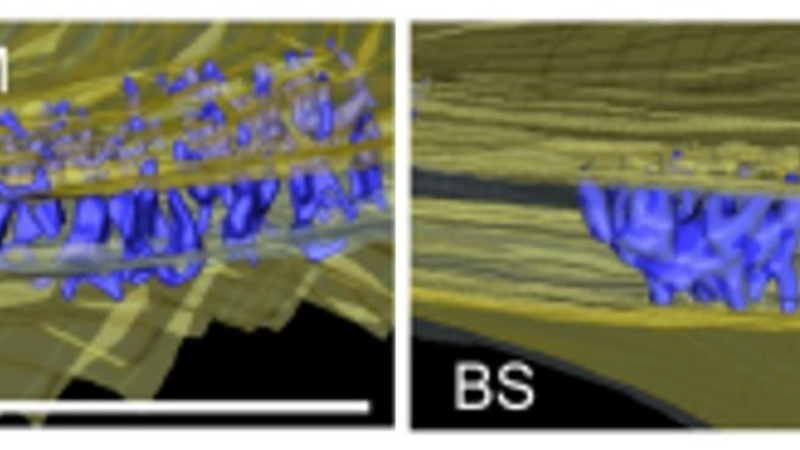
3D reconstructions of SBF-SEM image series illustrating plasmodesmal frequency at the mesophyll-bundle sheath cell interface in C4 plant G. gynandra (left) and the closely related C3 plant T. hassleriana (right). To see the full segmentation, 3D animation movies, please go to the supporting information here: https://doi.org/10.1111/nph.19343
